Create Custom conda Environment¶
Compute Resources
Have questions or need help with compute, including activation or issues? Follow this link.
storageN
The use of
storageNwithin these documents indicates that any storage platform can be used.- Current available storage platforms:
storage1
storage2
Docker Usage
The information on this page assumes that you have a knowledge base of using Docker to create images and push them to a repository for use. If you need to review that information, please see the links below.
Docker Basics: Building, Tagging, & Pushing A Custom Docker Image
Overview¶
The purpose of this tutorial is to demonstrate the steps required to build and use a custom conda-based environment in RIS using an existing Docker image. The conda environment will be located in the RIS Data Storage Platform so that any changes will persist between jobs.
As an example, a sequence analysis conda environment will be created containing the following tools:
citipy (https://pypi.org/project/citipy/)
fastqc (https://github.com/s-andrews/FastQC)
multiqc (https://multiqc.info/)
samtools (http://www.htslib.org/)
spades (https://cab.spbu.ru/software/spades/)
Interactive Job Submission¶
Defining Environment Variables¶
Begin by defining the environment variables that will be used during creation of
the conda environment. The environment variables can be defined in your
.bashrc or .condarc file to avoid having to enter them for each job.
The environment variables are:
CONDA_ENVS_DIRS: the path to the directory where conda environmentswill be created.
CONDA_PKGS_DIRS: the path to the directory where conda packageswill be downloaded to.
See the conda documentation for more information.
In the example below, the CONDA_ENVS_DIRS and CONDA_PKGS_DIRS are set to
a folder in the RIS Data Storage Platform. Make sure the folders exist and are
writable.
export CONDA_ENVS_DIRS="/storageN/fs1/${STORAGE_ALLOCATION}/Active/conda/envs/"
export CONDA_PKGS_DIRS="/storageN/fs1/${STORAGE_ALLOCATION}/Active/conda/pkgs/"
Please also define the following environment variables to mount the RIS Data
Storage Platform and add conda binaries to the PATH:
export LSF_DOCKER_VOLUMES="/storageN/fs1/${STORAGE_ALLOCATION}/Active:/storageN/fs1/${STORAGE_ALLOCATION}/Active"
export PATH="/opt/conda/bin:$PATH"
Job Submission¶
Once the appropriate environment variables are defined, the next step is to
submit the interactive job using the
continuumio/anaconda3:2021.11 Docker image.
bsub -Is -q general-interactive -a 'docker(continuumio/anaconda3:2021.11)' /bin/bash
Note
After the interactive job lands on an execution host, the command-line prompt
will begin with (base). For example:
(base) tahan@compute1-exec-132:~$
If this is not the case, conda needs to be initialized. Please see our documentation for instructions on how to resolve this issue.
Creating the conda Environment File¶
The conda environment will be created using an environment YAML file. The
interactive session has the nano text editor installed to create the file
with. The file will be saved in the home folder and named environment.yml.
Please see the conda documentation
for more information.
Note
For users wanting to create multiple environments, naming each YAML file
the same name as the environment is recommended to avoid confusion. If using
more than one YAML file, be sure to replace the occurrences of
environment.yml in the tutorial with the name of each YAML file.
Open a New File in nano named environment.yml:¶
nano ~/environment.yml
Copy and paste following into the file:
name: sequencing
channels:
- conda-forge
- bioconda
dependencies:
- bwa
- fastp
- fastqc
- multiqc>=1.7
- samtools<=1.11
- spades=3.9.1
- pip
- pip:
- citipy
Note
The pip dependency is required to install the citipy package using the
pip package manager. The pip dependencies are added to the end of the
environment file to reduce conda/pip installation issues from being used in
the same environment. This is the ideal order of dependencies and should be
followed to reduce possible installation issues.
The environment.yml file provides the following instructions for creating the conda environment:
name: the name of the conda environment. This name will be used to reference the environment in theconda activatecommand.channels: the channels to be used when creating the conda environment. Channels can be thought of as additional repositories that contain packages.dependencies: the packages that will be installed in the conda environment.pip: thepippackage manager is used to install additional packages. In the example, thecitipypackage is installed usingpip.Specific package versions can be specified using the
<,>and=operators.For example,
samtools<=1.11will install the latest version ofsamtoolsless than or equal to 1.11.
Please see the conda environment file documentation for more information.
Installing the conda Environment From environment.yml¶
Run the following command to install the conda environment using the
environment.yml file:
conda env create -f ~/environment.yml
The conda environment creation may take several minutes to complete. Please be patient as the environment is created. Once the environment is created, the following output will be displayed:
# To activate this environment, use
#
# $ conda activate sequencing
#
# To deactivate an active environment, use
#
# $ conda deactivate
Activating the conda Environment¶
Once the example sequencing conda environment is created, it can be
activated using the following command:
conda activate sequencing
All of the packages listed in the environment.yml file are available for
use. To view a list of the installed packages in an environment, run the
following command after activating the environment:
conda list
For example, the sequencing environment will list the following packages:
(sequencing) tahan@compute1-exec-132:~$ conda list
# packages in environment at /storageN/fs1/tahan/Active/projects/conda/envs/sequencing:
#
# Name Version Build Channel
_libgcc_mutex 0.1 conda_forge conda-forge
_openmp_mutex 4.5 1_llvm conda-forge
blas 2.17 openblas conda-forge
bwa 0.7.17 h5bf99c6_8 bioconda
bzip2 1.0.8 h7f98852_4 conda-forge
c-ares 1.18.1 h7f98852_0 conda-forge
ca-certificates 2021.10.8 ha878542_0 conda-forge
certifi 2018.8.24 py35_1001 conda-forge
citipy 0.0.5 pypi_0 pypi
click 7.1.2 pyh9f0ad1d_0 conda-forge
colormath 3.0.0 py_2 conda-forge
cycler 0.10.0 py_2 conda-forge
dbus 1.13.6 h48d8840_2 conda-forge
decorator 5.1.1 pyhd8ed1ab_0 conda-forge
expat 2.4.3 h9c3ff4c_0 conda-forge
fastp 0.23.2 h79da9fb_0 bioconda
fastqc 0.11.9 hdfd78af_1 bioconda
font-ttf-dejavu-sans-mono 2.37 hab24e00_0 conda-forge
fontconfig 2.13.1 he4413a7_1000 conda-forge
freetype 2.10.4 h0708190_1 conda-forge
future 0.16.0 py35_2 conda-forge
gettext 0.19.8.1 h0b5b191_1005 conda-forge
glib 2.68.4 h9c3ff4c_0 conda-forge
glib-tools 2.68.4 h9c3ff4c_0 conda-forge
gst-plugins-base 1.14.0 hbbd80ab_1
gstreamer 1.14.0 h28cd5cc_2
htslib 1.11 hd3b49d5_2 bioconda
icu 58.2 hf484d3e_1000 conda-forge
importlib-metadata 2.0.0 py_1 conda-forge
isa-l 2.30.0 ha770c72_4 conda-forge
jinja2 2.11.3 pyh44b312d_0 conda-forge
jpeg 9d h36c2ea0_0 conda-forge
kdtree 0.16 pypi_0 pypi
kiwisolver 1.0.1 py35h2d50403_2 conda-forge
krb5 1.19.2 hcc1bbae_3 conda-forge
libblas 3.8.0 17_openblas conda-forge
libcblas 3.8.0 17_openblas conda-forge
libcurl 7.81.0 h2574ce0_0 conda-forge
libdeflate 1.7 h7f98852_5 conda-forge
libedit 3.1.20191231 he28a2e2_2 conda-forge
libev 4.33 h516909a_1 conda-forge
libffi 3.3 h58526e2_2 conda-forge
libgcc-ng 11.2.0 h1d223b6_11 conda-forge
libgfortran-ng 7.5.0 h14aa051_19 conda-forge
libgfortran4 7.5.0 h14aa051_19 conda-forge
libglib 2.68.4 h3e27bee_0 conda-forge
libiconv 1.16 h516909a_0 conda-forge
liblapack 3.8.0 17_openblas conda-forge
liblapacke 3.8.0 17_openblas conda-forge
libnghttp2 1.43.0 h812cca2_1 conda-forge
libopenblas 0.3.10 pthreads_hb3c22a3_5 conda-forge
libpng 1.6.37 h21135ba_2 conda-forge
libssh2 1.10.0 ha56f1ee_2 conda-forge
libstdcxx-ng 11.2.0 he4da1e4_11 conda-forge
libuuid 2.32.1 h7f98852_1000 conda-forge
libxcb 1.13 h7f98852_1004 conda-forge
libxml2 2.9.9 h13577e0_2 conda-forge
libzlib 1.2.11 h36c2ea0_1013 conda-forge
llvm-openmp 12.0.1 h4bd325d_1 conda-forge
lzstring 1.0.4 py_1001 conda-forge
markdown 3.3.3 pyh9f0ad1d_0 conda-forge
markupsafe 1.0 py35h470a237_1 conda-forge
matplotlib 3.0.0 py35h5429711_0
more-itertools 8.12.0 pyhd8ed1ab_0 conda-forge
multiqc 1.7 py_4 bioconda
ncurses 6.2 h58526e2_4 conda-forge
networkx 2.4 py_1 conda-forge
numpy 1.15.2 py35h99e49ec_0
numpy-base 1.15.2 py35h2f8d375_0
openjdk 11.0.1 h516909a_1016 conda-forge
openssl 1.1.1l h7f98852_0 conda-forge
pcre 8.45 h9c3ff4c_0 conda-forge
perl 5.32.1 1_h7f98852_perl5 conda-forge
pip 20.3.4 pyhd8ed1ab_0 conda-forge
pthread-stubs 0.4 h36c2ea0_1001 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.9.2 py35h05f1152_2
python 3.5.6 h12debd9_1
python-dateutil 2.8.1 py_0 conda-forge
pytz 2021.3 pyhd8ed1ab_0 conda-forge
pyyaml 3.12 py35_1 conda-forge
qt 5.9.7 h5867ecd_1
readline 8.1 h46c0cb4_0 conda-forge
requests 2.13.0 py35_0 conda-forge
samtools 1.11 h6270b1f_0 bioconda
setuptools 40.4.3 py35_0 conda-forge
simplejson 3.16.1 py35h470a237_0 conda-forge
sip 4.19.8 py35hf484d3e_1000 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
spades 3.9.1 h9ee0642_1 bioconda
spectra 0.0.11 py_1 conda-forge
sqlite 3.37.0 h9cd32fc_0 conda-forge
tk 8.6.11 h27826a3_1 conda-forge
tornado 5.1.1 py35h470a237_0 conda-forge
wheel 0.37.1 pyhd8ed1ab_0 conda-forge
xorg-libxau 1.0.9 h7f98852_0 conda-forge
xorg-libxdmcp 1.1.3 h7f98852_0 conda-forge
xz 5.2.5 h516909a_1 conda-forge
yaml 0.2.5 h7f98852_2 conda-forge
zipp 1.0.0 py_0 conda-forge
zlib 1.2.11 h36c2ea0_1013 conda-forge
Using the above command, we can ensure that the version requirements for the conda environment were met. As a reminder, the environment file had the following version requirements:
multiqc>=1.7
samtools<=1.11
spades=3.9.1
From the package list, we can validate that the version requirements were met.
# Name Version Build Channel
multiqc 1.7 py_4 bioconda
samtools 1.11 h6270b1f_0 bioconda
spades 3.9.1 h9ee0642_1 bioconda
A list of the currently installed environments can be viewed with the command:
conda env list
Compatible Docker Images¶
The following Docker images have been tested with this tutorial to create a custom conda environment:
Jupyter Notebook Data Science Stack (https://hub.docker.com/r/jupyter/datascience-notebook/).
Tested with
jupyter/datascience-notebook:ubuntu-20.04
mambaforge (https://hub.docker.com/r/condaforge/mambaforge).
Tested with
condaforge/mambaforge:4.11.0-0Replace
condacommands withmamba. See the mamba documentation for more information.
Creating IPython Kernel for Usage with Jupyter Notebooks/Labs¶
In order to create an IPython Kernel you will need to first activate your conda environment that you created (using the above examples) and then follow the below commands:
PATH=/opt/conda/bin:PATH bsub -Is -q general-interactive -a 'docker(continuumio/anaconda3:2021.11)' /bin/bash
conda init #initializes shell
source .bashrc #activates base conda environment
conda env list #Lists all conda environment
conda activate "env_name"
Then to install IPython Kernel you would need run the below commands to install the needed IPython packages and then register your kernel for this particular conda environment:
pip install ipython
python -m pip install ipykernel
python -m ipykernel install --user --name "Some_Name" --display-name "User_Friendly_Name"
Note
Make sure to replace “env_name”, “Some_Name” and “User_Friendly_Name” with respected Values.
Then once you have a Jupyter Notebook/Lab up and running on RIS platform (through OnDemand Portal or Command Line). Select the Kernel using either the top right corner button or the Kernel menu in the Menu bar.
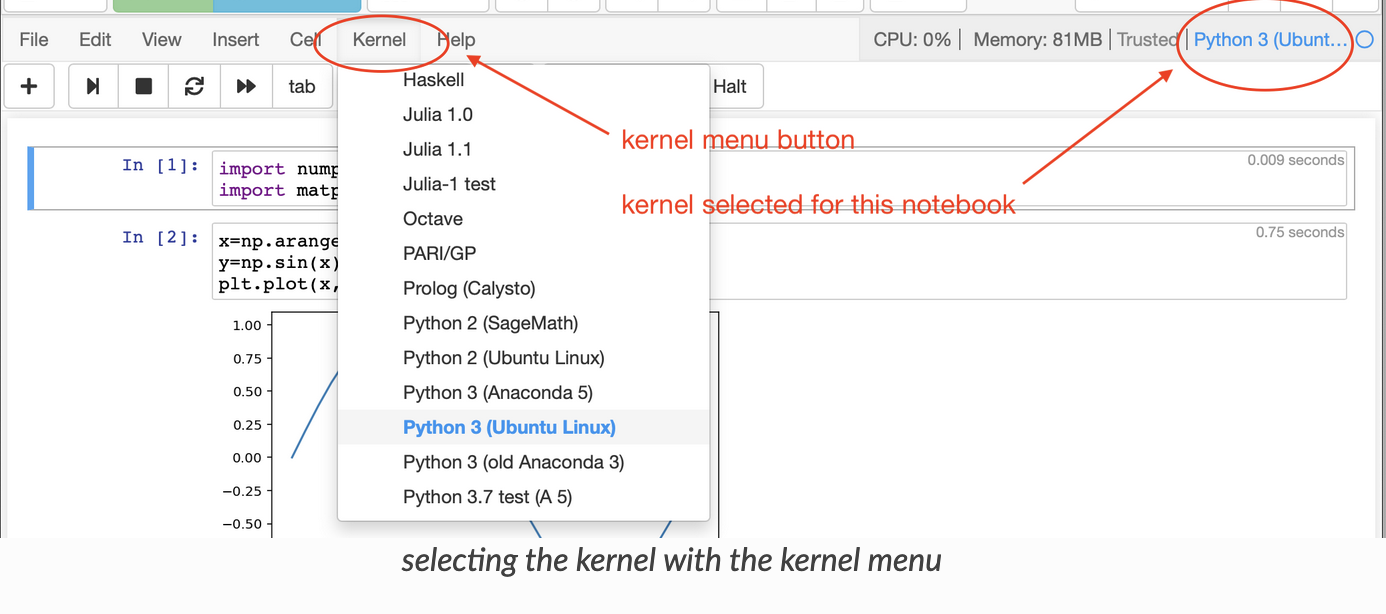
Note
You will need to repeat these steps for each conda environment for which you wish to register a kernel and use with a Jupyter Notebook/Lab.
